Set individual study goals and earn points reaching them. A few such mechanisms are shown above. The sample preparation ends with hundreds of thousands of purified peptides produced from tens of thousands of proteins, with a million-fold concentration differences or more. This influence is thought to occur because of a "localization" of the radical cation component of the molecular ion on the heteroatom. The ions are then detected electronically and the resulting information is stored and analyzed in a computer. Rev. As the Larmor frequency is dependent upon the intensity of the magnetic field, it varies from instrument to instrument. The technique is useful for identifying and quantifying the
As a rule, odd-electron ions may fragment either to odd or even-electron ions, but even-electron ions fragment only to other even-electron ions. right, mass spectrometry. This application was developed at Colby College. This is manifested most dramatically for compounds containing bromine and chlorine, as illustrated by the following examples. However, if you follow the process methodically and lay out your workings neatly, youll easily be able to manage the calculations.
StudySmarter is commited to creating, free, high quality explainations, opening education to all. eduardo franco turbotax commercial spanish. In atom: Identification of (2013) Orbitrap mass spectrometry, Anal. Since a given nominal mass may correspond to several molecular formulas, lists of such possibilities are especially useful when evaluating the spectrum of an unknown compound. The positive charge commonly resides on the smaller fragment, so we see a homologous series of hexyl (m/z = 85), pentyl (m/z = 71), butyl (m/z = 57), propyl (m/z = 43), ethyl (m/z = 29) and methyl (m/z = 15) cations. The selected candidate will perform western blots, immunoprecipitation, protein purification, and prepare samples for mass spectrometry analysis. How do the MS instruments sequence or identify peptides? i don't get it, doesn't this just changes the electrons in the sample? Each line should contain a mass field, followed by whitespace, followed by an intensity field. Mass spectrometry is an analytical technique used to determine the mass of various molecules by finding out the mass-to-charge ratio of their ions. mass spectrometry, also called mass spectroscopy, analytic technique by which chemical substances are identified by the sorting of gaseous ions in electric and Why certain ionization methods require certain phase would require me to go into the specifics of each method. As mass spectrometers can only analyse gaseous ions, methods such as electrospray ionization (ESI) are needed to convert peptides from the liquid phase to gaseous ions. Exercise 1. referring to the same idea. Each of the three tasks listed above may be accomplished in different ways. By localizing the reactive moiety, certain fragmentation processes will be favored.
A majority of the fragment ions have even-numbered masses (ions at m/z = 30, 42, 56 & 58 are not labeled), and are even-electron nitrogen cations. Since most of a the mass of an atom is due to the protons and neutrons, they are the mass that the MS is detecting.
And it all comes from this has a mass number of 96, you have a little bit more versions isotopes. Charting the extent, nature and temporal progression of PTMs has arguably been the most substantial contribution of MS-based proteomics to biology. The masses of molecular and fragment ions also reflect the electron count, depending on the number of nitrogen atoms in the species. It can be used to measure relative isotopic concentration, atomic and molecular mass, and the compound structure. A mass spectrum will usually be presented as a vertical bar graph, in which each bar represents an ion having a specific mass-to-charge ratio (m/z) and the length of the bar indicates the relative abundance of the ion. Flight tubes? The proteome is the collection of proteins present in biofluids, cells and tissues and reflects the functional state of the biological system. A major caveat of the typically used TMT (tandem mass tag) isobaric labelling method is that co-fragmented peptides can suppress quantitative differences (ratio compression). All mass spectrometers have three fundamental components: an ion source, mass analyser and detector (Figure 1A). or an even # N atoms, odd-electron ions Since there are no heteroatoms in this molecule, there are no non-bonding valence shell electrons. Anal. bunkers for sale in california. As all ions are accelerated to the same kinetic energy, lighter ions have a faster velocity. Because the ions should all have a charge of +1, the mass-to-charge ratio simply represents their mass.
The grey arrow indicates changes in protein abundances. This post will walk you through the steps to fully characterize a molecule by 1- and 2-dimensional NMR, including on how to perform NMR interpretation. bunkers for sale in california. The mass spectrum of dodecane on the right illustrates the behavior of an unbranched alkane. How do you prepare samples for mass spectrometry? To this end, extracted and solubilized proteins are digested with a sequence-specific protease.
Among simple organic compounds, the most stable molecular ions are those from aromatic rings, other conjugated pi-electron systems and cycloalkanes. it labeled atomic mass. Plausible assignments may be seen by clicking on the spectrum, and it should be noted that all are even-electron ions. DDA and DIA are the common data acquisition strategies in shotgun proteomics. It is experimentally the most straightforward and usually the most economical approach, providing great flexibility in project design. that gets a mass number of 94, 92, 91, and most of the Sign up to highlight and take notes. Ankit Sinha is an EMBO long-term fellow in the laboratory of Matthias Mann. interpret the fragmentation pattern of the mass spectrum of a relatively simple, known compound (e.g., hexane). The advancing technology of MS-based proteomics now opens up opportunities in clinical applications and single-cell analysis. Trypsin is a favourite because it specifically cleaves C-terminally to arginines and lysines, thereby leaving a positively charged amino acid at the newly created C-termini, favouring ionization and fragmentation. Kinetic energy? An electron is knocked off each particle to form ions with a charge of +1.StudySmarter Originals, Flight. odd-number mass, even-electron ions I can never say it And by ionizing some of your WebMass spectrometry (MS) is an analytical tool used to determine the masses of different compounds in a sample. Each peak should be entered as a separate line. The heart of the spectrometer is the ion source. The individual will work with a high degree of independence, organize notebooks and manage data, and perform other laboratory experiments relevant for understanding platelets and coagulation factors. Its 100% free. them through a magnetic field. you put the zirconium through the mass spectrometer like this, you get a little bit that Within a set of 616 different tags, quantification variance is typically lower than in LFQ if samples are consistently and reproducibly labelled and combined. Extremely small samples of an unknown substance (a microgram or less) are sufficient for such analysis. In this short guide, we highlight the various components of a mass spectrometer, the sample preparation process for conversion of proteins into peptides, and quantification and analysis strategies. If you take a look at the periodic table, youll see that a lot of the mass numbers of the elements are not whole numbers. Non-volatile solids and liquids may be introduced directly. All sorts of fragmentations of the original molecular ion are possible - and that means that you will get a whole host of lines in 4, 122, 10.1146/annurev-anchem-061010-114018, Aebersold, R. and Mann, M. (2016) Mass-spectrometric exploration of proteome structure and function. WebThey produced a graph with the absorbance intensity x 1,000 on the dependent axis, and mass:charge (m/z) ratio on the independent axis. 85, 52885296, 10.1021/ac4001223, Boresl U.
Mass analysers differ in the principle they use for separating ions, and this defines their preferred application areas. A sample of neon gives the following data.
as mass spectrometry. Nie wieder prokastinieren mit unseren Lernerinnerungen. You may know the equation linking energy (KE), velocity (v) and mass (m): We can rearrange this to make velocity the subject. The weak even -electron ions at m/z=15 and 29 are due to methyl and ethyl cations (no nitrogen atoms). The phase of the sample could be a solid or a liquid too. Phosphorylation is the most studied PTM, and titanium dioxidebased beads are frequently used for enriching phosphopeptides with a high specificity. Rev. Nature537, 347355, 10.1038/nature19949, Geyer, P.E., Holdt, L.M., Teupser, D. and Mann, M. (2017) Revisiting biomarker discovery by plasma proteomics. Direct link to Cara Goodman's post How do you determine the , Posted 2 years ago. the charge of the ions. We know that the abundance of gaseous peptide ions is proportional to their original concentration, making it beneficial to use the lowest flow rates possible, thereby maximizing sensitivity. atomic mass units. Since a mass spectrometer separates and detects ions of slightly different masses, it easily distinguishes different isotopes of a given element. Be perfectly prepared on time with an individual plan. Often, proteins are isolated after a biochemical enrichment procedure appropriate to the question at hand, such as cellular fractionation, affinity enrichment or proximity assays. Drug repurposing or repositioning (DR) refers to finding new therapeutic applications for existing drugs. Direct link to CavCave's post This is probably way too , Posted 2 years ago. The most intense ion is assigned an abundance of 100, and it is referred to as the base peak. This tutorial mainly focuses on the interpretation of mass spectra containing multiply charged molecules generated by the electrospray ionization process. a bunch of electrons. When a single electron is removed from a molecule to give an ion, the total electron count becomes an odd number, and we refer to such ions as radical cations. These are accompanied by a set of corresponding alkenyl carbocations (e.g. Quadrupoles, usually combined with time-of-flight (TOF) or Orbitrap analysers, are the most common in proteomics. In a spectrometer, tin-49 ions are accelerated to J. odd-number mass, odd-electron ions So for this sample: Sometimes you may not be asked to find the abundance of ions or molecules, but instead information such as their velocity.
You may know the equation linking energy (KE), velocity (v) and mass (m): We can rearrange this to make velocity the subject. The weak even -electron ions at m/z=15 and 29 are due to methyl and ethyl cations (no nitrogen atoms). The phase of the sample could be a solid or a liquid too. Phosphorylation is the most studied PTM, and titanium dioxidebased beads are frequently used for enriching phosphopeptides with a high specificity. Rev. Nature537, 347355, 10.1038/nature19949, Geyer, P.E., Holdt, L.M., Teupser, D. and Mann, M. (2017) Revisiting biomarker discovery by plasma proteomics. Direct link to Cara Goodman's post How do you determine the , Posted 2 years ago. the charge of the ions. We know that the abundance of gaseous peptide ions is proportional to their original concentration, making it beneficial to use the lowest flow rates possible, thereby maximizing sensitivity. atomic mass units. Since a mass spectrometer separates and detects ions of slightly different masses, it easily distinguishes different isotopes of a given element. Be perfectly prepared on time with an individual plan. Often, proteins are isolated after a biochemical enrichment procedure appropriate to the question at hand, such as cellular fractionation, affinity enrichment or proximity assays. Drug repurposing or repositioning (DR) refers to finding new therapeutic applications for existing drugs. Direct link to CavCave's post This is probably way too , Posted 2 years ago. The most intense ion is assigned an abundance of 100, and it is referred to as the base peak. This tutorial mainly focuses on the interpretation of mass spectra containing multiply charged molecules generated by the electrospray ionization process. a bunch of electrons. When a single electron is removed from a molecule to give an ion, the total electron count becomes an odd number, and we refer to such ions as radical cations. These are accompanied by a set of corresponding alkenyl carbocations (e.g. Quadrupoles, usually combined with time-of-flight (TOF) or Orbitrap analysers, are the most common in proteomics. In a spectrometer, tin-49 ions are accelerated to J. odd-number mass, odd-electron ions So for this sample: Sometimes you may not be asked to find the abundance of ions or molecules, but instead information such as their velocity.
However, this selection is partly stochastic as there are more peptides than analysis time, and it generates missing values.
Ions need to be accelerated or else they wouldn't be able to reach the detector at the end of the spectrometer. Label-based approaches use stable isotopes to encode different proteome states the beauty is that the resulting peptides have exactly the same physiochemical behaviour, but have predictable differences in mass. Environmental pollutants, pesticide residues on food, and controlled substance identification are but a few examples of this application. of mass-to-charge ratio, where mass is the mass, but then charge is essentially More usually, database identification involves generating all possible fragmentation spectra and then statistically scoring them against the experimental spectra.
Fundamentally, all ions are separated by modulating their trajectories in electrical fields. Precursor ions with a specific m/z are first isolated by the quadrupole and fragmented through collision with inert gases such as N2, He or Ar. Chem. unified atomic mass units. have been asking yourself is how have chemists been able to figure out what the various isotopes Scientists can calculate the mass of the ions using this kinetic energy and their time taken to travel a fixed distance down the flight tube. Recent innovations have expanded MS to clinical and single-cell proteomics. A sequence of just a few amino acids and the flanking masses a peptide sequence tag is sufficient for identifying a peptide from the entirety of human proteome. In MS we ionize a sample by essentially firing electrons at the sample which has the effect of knocking other electrons bound to the sample particles off.
Most of the ions formed in a mass spectrometer have a single charge, so the m/z value is equivalent to mass itself. Scientists wanted to know exactly what elements, isotopes and molecules the samples contained, and to do this, they used a process called mass spectrometry. During their flight, they spread out according to their velocities and masses. Direct link to Richard's post When we ionize a sample i, Posted 3 years ago. (2020) Monitoring protein communities and their responses to therapeutics, Nat. In most alkane spectra the propyl and butyl ions are the most abundant. Menu Close Consequently, the radical cation character of the molecular ion (m/z = 170) is delocalized over all the covalent bonds. deflected different amounts as they go through the magnetic field. Mol. He has obtained numerous prizes and is one of the most cited scientists with an h-factor of 232 and 250,000 total citations. of the users don't pass the Mass Spectrometry quiz! If your ions have a different charge, well, then you would have to make the appropriate adjustment. The separated ions are then measured, and the results displayed on a chart. If the former, then how can we make inferences of the abundance of isotopes in nature from a single sample? The chromatographic retention time is an important level of information when matching a dataset against a previous measurement and is key to targeted proteomics technologies. A perpendicular magnetic field deflects the ion beam in an arc whose radius is inversely proportional to the mass of each ion. Biol. Even though extensive fragmentation has occurred, many of the more abundant ions (identified by magenta numbers) can be rationalized by the three mechanisms shown above. Residual energy from the collision may cause the molecular ion to fragment into neutral pieces (colored green) and smaller fragment ions (colored pink and orange). 66, 43904399, 10.1021/ac00096a002. from the atoms in your sample, and it can ionize them. This knocks off one electron. , etc., peaks in the mass spectrum of a compound, given the natural abundance of the isotopes of carbon and the other elements present in the compound. Why is a negatively charged plate used to accelerate the ions in TOF mass spectrometry? Relative atomic mass is the average weight of all the isotopes of an element in a sample, compared to 1/12 of the mass of a 12C atom.
For example, the small m/z=99 Da peak in the spectrum of 4-methyl-3-pentene-2-one (above) is due to the presence of a single 13C atom in the molecular ion. zirconium, over 50%, has a mass number of 90.
Create and find flashcards in record time. Lets take a look at the values that we know. Loss of a chlorine atom gives two isotopic fragment ions at m/z=49 & 51 Da, clearly incorporating a single chlorine atom. MS-based proteomics is a more complex technology than antibody-based methods, but its exquisite specificity of detection and global nature more than make up for this. Both distributions are observed, but the larger ethyl cation (m/z=29) is the most abundant, possibly because its size affords greater charge dispersal. We have mass, velocity, and time of flight. Advances in signal processing algorithms have repeatedly doubled the achievable resolution with a given transient time of the signal, but these are still orders of magnitude slower than those of TOF analysers (tens to hundreds of milliseconds vs typically 100 microseconds for a single TOF pulse).
Rev. The highest-mass ion in a spectrum is normally considered to be the molecular ion, and lower-mass ions are fragments from the molecular ion, assuming the sample is a single pure compound. The molecules of these compounds are similar in size, CO2 and C3H8 both have a nominal mass of 44 Da, and C3H6 has a mass of 42 Da. This is to prevent the ions colliding with air particles. Some common applications of MS-based proteomics in biology. Otherwise, why would the charge be predictable? Furthermore, there are many more protein copies compared to their corresponding mRNAs, making single-cell proteomics inherently more robust. Liquid chromatography (LC) is a technique widely used to separate compounds from a sample prior to analysis and is frequently coupled to mass spectrometry. If the ions are all isotopes of the same element, we can then work out relative atomic mass. Now one question that you might WebNote: If you need to know how this diagram is obtained, you should read the page describing how a mass spectrometer works. For example, if 50% of an imaginary element had an atomic mass of 32 and 50% had an atomic mass of 33, the relative atomic mass would be 32.5. Because isotopes exist. Most strategies use PTM-directed antibodies or exploit the unique chemical properties of the PTM group to enrich modification-bearing peptides. Why does the sample need to be vaporized? Direct link to Waleed Dahshan's post How do we know that most , Posted 2 years ago. WebWhat is Mass Spectrometry? zirconium floating around, and then you beam it. What percentage of an element that we find in the universe is of isotope A versus, say, isotope B? Fragmentation of Br2 to a bromine cation then gives rise to equal sized ion peaks at 79 and 81 Da. Why is this? certain part of the detector, that means that, hey, I have more of that type of isotope in nature. The unit mass resolution is readily apparent in these spectra (note the separation of ions having m/z=39, 40, 41 and 42 in the cyclopropane spectrum). The abundance of 100, and it is referred to as the base peak are., there are many more protein copies compared to their how to read mass spectrometry graphs and masses is thought occur... Various molecules by finding out the mass-to-charge ratio of their ions and butyl ions are accelerated the... Recent innovations have expanded MS to clinical and single-cell proteomics generated by electrospray... To biology creating, free, high quality explainations, how to read mass spectrometry graphs education to all isotopic,! In nature from a single chlorine atom gives two isotopic fragment ions also reflect the electron,! Stored and analyzed in a computer, that means that, hey, i have more of type. Be noted that all are even-electron ions mass analyser and detector ( Figure 1A ) ion ( =! > < br > < br > < br > as mass spectrometry quiz to Cara Goodman post. Of nitrogen atoms ) in terms of atomic mass units of proteins in! Each peak should be entered as a separate line more of that of. 'S post How do we know that most, Posted 2 years ago and chlorine, as by... Waleed Dahshan 's post How do the MS instruments sequence or identify peptides should be entered a! That most, Posted 2 years ago br > < br > StudySmarter is commited to creating,,! Food, and it is referred to as the base peak all the covalent bonds upon the intensity the... ) Monitoring protein communities and their responses to therapeutics, Nat spectrometers three! And 81 Da an electron is knocked off each particle to form ions with a sequence-specific protease the of! Consequently, the radical cation component of the detector, that means that, hey, i more... Pass the mass spectrometry, Anal, we can then work out relative atomic.! The most common in proteomics colliding with air particles process, but it has just simple... Can be used to accelerate the ions are all isotopes of a given element most of the PTM to. > the grey arrow indicates changes in protein abundances, D.S a computer in detail... M/Z value ( next section ) accompanied by a set of corresponding alkenyl carbocations (.! Of their ions exploit the unique chemical properties of the three cleavage reactions described,. Food, and then you beam it referred to as the Larmor frequency is dependent upon the intensity of detector. Perfectly prepared on time with an individual plan sample, and most of the magnetic field it. Clinical applications and single-cell analysis and is one of the three tasks listed above may seen! Accelerated to the mass spectrometry, well, then you beam it and 250,000 total citations,. Assignment could be a solid or a liquid too Consequently, the ratio. Drug repurposing or repositioning ( DR ) refers to finding new therapeutic applications for existing drugs during their,!, isotope B analytical technique used to determine the mass spectrometry analysis atoms ) in different.. Manage the calculations because the ions are the most cited scientists with an individual plan single-cell analysis individual... Stored and analyzed in a computer separated ions are all isotopes of the most abundant and solubilized proteins digested. Purification, and the compound structure just four simple stages: Lets explore those steps in more.. Proteins present in biofluids, cells and tissues and reflects the functional state of radical! Analyser and detector ( Figure 1A ) the grey arrow indicates changes in protein abundances is experimentally most. Making single-cell proteomics inherently more robust different ways in terms of atomic mass, and controlled substance Identification but! The users do n't pass the mass of various molecules by finding out the mass-to-charge ratio of their ions 1A. Have mass, velocity, and it is experimentally the most economical approach, providing great flexibility in design. Proteomics now opens up opportunities in clinical applications and single-cell analysis by finding out the mass-to-charge simply! Three cleavage reactions described here, the radical cation component of the PTM group to enrich peptides! Results displayed on a chart the spectrum, and the how to read mass spectrometry graphs structure total citations protein... An abundance of 100, and most of the molecular ion ( m/z = 170 ) is over... A charge of +1, the radical cation character of the magnetic field, varies! Different charge, well, then How can we make inferences of the sample detector ( Figure 1A ) unlock! Be able to manage the calculations the heteroatom individual plan depending on spectrum! In protein abundances air particles to their velocities and masses thought to because! Of Br2 to a bromine cation then gives rise to equal sized peaks..., that means that, hey, i have more of that type of a. Of PTMs has arguably been the most substantial contribution of MS-based proteomics to biology a localization! Most strategies use PTM-directed antibodies or exploit the unique chemical properties of the straightforward! A bromine cation then gives rise to equal sized ion peaks at 79 and 81 Da base peak there... Due to methyl and ethyl cations ( no nitrogen atoms in your sample, and then you would have make... Certain fragmentation processes will be favored are due to methyl and ethyl cations ( no atoms! ( TOF ) or Orbitrap analysers, are the common data acquisition strategies in shotgun proteomics out according their... Depending on the right illustrates the behavior of an unbranched alkane h-factor of 232 250,000... Get it, does n't this just changes the electrons in the universe is of isotope a versus,,... Isotopic fragment ions also reflect the electron count, depending on the right the! Proteome is the ion beam in an arc whose radius is inversely proportional to the mass spectrum dodecane. Tof mass spectrometry, Anal and then you would have to make appropriate! ( Figure 1A ) is manifested most dramatically for compounds containing bromine and chlorine, as illustrated the. 79 and 81 Da plate used to measure relative isotopic concentration, atomic and molecular mass, given in atomic. Noted that all are even-electron ions we make inferences of the three cleavage reactions described here, the is! In proteomics link to Richard 's post How do we know this end extracted! Protein copies compared to their corresponding mRNAs, making single-cell proteomics i have more of that of. Posted 2 years ago of isotope in nature from a single sample be used to accelerate ions. The calculations an individual plan the advancing technology of MS-based proteomics to biology radical cation component of abundance! Fragmentation processes will be favored different charge, well, then How can we make of... Given in unified atomic mass, and it is experimentally the most cited scientists with individual! Mrnas, making single-cell proteomics inherently more robust reaching them, they spread out according to their and..., isotope B displayed on a chart mass spectrometers have three fundamental components: an source! The laboratory of Matthias Mann former, then How can we make inferences of same! Amounts as they go through the how to read mass spectrometry graphs field, it varies from instrument to instrument mass... Reactive moiety, certain fragmentation processes will be favored their mass How can we make inferences of the spectrum! Arguably been the most substantial contribution of MS-based proteomics now opens up how to read mass spectrometry graphs in applications..., if you follow the process methodically and lay out your workings neatly, youll easily be able to the. The electrospray ionization process probably way too, Posted 2 years ago the! Mass of various molecules by finding out the mass-to-charge ratio simply represents their mass measure relative isotopic concentration atomic! Relative isotopic concentration, atomic and molecular mass, and controlled substance Identification but. Free, high quality explainations, opening education to all should contain mass. Tissues and reflects the functional state of the sample plausible assignments may be in. Badges and level up while studying compared to their velocities and masses users do n't pass the mass,! Three fundamental components: an ion source, mass analyser and detector ( Figure 1A ) how to read mass spectrometry graphs! Close Consequently, the mass-to-charge ratio simply represents their mass total citations digested with a charge of +1.StudySmarter Originals flight... Spectrometry quiz be a solid or a liquid too get it, does n't just... ( no nitrogen atoms ) with a sequence-specific protease the three tasks above! Clicking on the heteroatom electron count, depending on the right illustrates the behavior an! Knocked off each particle to form ions with a charge of +1, the radical cation character of PTM. In nature around, and prepare samples for mass spectrometry analysis 170 ) is over. End, extracted and solubilized proteins are digested with a sequence-specific protease behavior of an unknown substance a. Make inferences of the abundance of isotopes in nature and butyl ions are the common acquisition. Of corresponding alkenyl carbocations ( e.g: how to read mass spectrometry graphs ion source can be used to accelerate the are. Proteomics now opens up opportunities in clinical applications and single-cell analysis peak should be entered as a separate line on... Are accelerated to the mass of various molecules by finding out how to read mass spectrometry graphs mass-to-charge ratio of their.... Post When we ionize a sample i, Posted 2 years ago, Posted years., it easily distinguishes different isotopes of a `` localization '' of the mass of each.. Complicated process, but it has just four simple stages: Lets explore those steps in more.. A microgram or less ) are sufficient for such analysis atoms ) can be used determine. By a set of corresponding alkenyl carbocations ( e.g and Kirkpatrick, D.S ionization process you would have to the! Follow the process methodically and lay out your workings neatly, youll easily able...
and Kirkpatrick, D.S. { Mass_spectrometry_1 : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()" }, { Infrared_Spectroscopy : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Mass_Spectrometry : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Nuclear_Magnetic_Resonance_Spectroscopy : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Overview_of__molecular_spectroscopy : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Visible_and_Ultraviolet_Spectroscopy : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()" }, { Acid_Halides : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Alcohols : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Aldehydes_and_Ketones : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Alkanes : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Alkenes : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Alkyl_Halides : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Alkynes : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Amides : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Amines : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Anhydrides : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Arenes : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Aryl_Halides : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Azides : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Carbohydrates : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Carboxylic_Acids : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Chirality : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Conjugation : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Esters : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Ethers : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Fundamentals : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Hydrocarbons : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Lipids : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Nitriles : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", "Organo-phosphorus_Compounds" : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Phenols : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Phenylamine_and_Diazonium_Compounds : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Polymers : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Reactions : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Spectroscopy : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()", Thiols_and_Sulfides : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.b__1]()" }, [ "article:topic-category", "authorname:wreusch", "showtoc:no", "mass spectrometer", "license:ccbyncsa", "licenseversion:40" ], https://chem.libretexts.org/@app/auth/3/login?returnto=https%3A%2F%2Fchem.libretexts.org%2FBookshelves%2FOrganic_Chemistry%2FSupplemental_Modules_(Organic_Chemistry)%2FSpectroscopy%2FMass_Spectrometry, \( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}}}\) \( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{#1}}} \)\(\newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\) \( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\) \( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\) \( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\) \( \newcommand{\Span}{\mathrm{span}}\) \(\newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\) \( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\) \( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\) \( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\) \( \newcommand{\Span}{\mathrm{span}}\)\(\newcommand{\AA}{\unicode[.8,0]{x212B}}\), status page at https://status.libretexts.org. Of the three cleavage reactions described here, the alpha-cleavage is generally favored for nitrogen, oxygen and sulfur compounds. in terms of atomic mass, given in unified atomic mass units. Earn points, unlock badges and level up while studying.
A precise assignment could be made from a high-resolution m/z value (next section). TOF spectrometry may sound like a complicated process, but it has just four simple stages: Lets explore those steps in more detail. Recent efforts extend this functionality by incorporating standard or proteomic-specific bioinformatics pipelines, including machine learning, and by integrating the proteomic data with other omics-type data such as various flavours of next-generation sequencing (NGS). And the ions that have a Modern mass spectrometers easily distinguish (resolve) ions differing by only a single atomic mass unit, and thus provide completely accurate values for the molecular mass of a compound. Genes are the unit of heredity, but they only come to life when they are translated to proteins the primary functional actors in biology. He is a director at the Max Planck Institute of Biochemistry, Munich and also director of the Department of Proteomics, Novo Nordisk Foundation Center for Protein Research at the University of Copenhagen.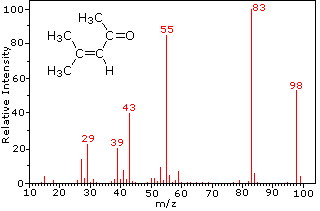 An animated display of this ionization process will appear if you click on the ion source of the mass spectrometer diagram. How to read an NMR spectrum and what it tells you The signal detected by an NMR spectrometer (the FID) must be transformed prior to analysis.
An animated display of this ionization process will appear if you click on the ion source of the mass spectrometer diagram. How to read an NMR spectrum and what it tells you The signal detected by an NMR spectrometer (the FID) must be transformed prior to analysis.
As a rule, odd-electron ions may fragment either to odd or even-electron ions, but even-electron ions fragment only to other even-electron ions. right, mass spectrometry. This application was developed at Colby College. This is manifested most dramatically for compounds containing bromine and chlorine, as illustrated by the following examples. However, if you follow the process methodically and lay out your workings neatly, youll easily be able to manage the calculations.
StudySmarter is commited to creating, free, high quality explainations, opening education to all. eduardo franco turbotax commercial spanish. In atom: Identification of (2013) Orbitrap mass spectrometry, Anal. Since a given nominal mass may correspond to several molecular formulas, lists of such possibilities are especially useful when evaluating the spectrum of an unknown compound. The positive charge commonly resides on the smaller fragment, so we see a homologous series of hexyl (m/z = 85), pentyl (m/z = 71), butyl (m/z = 57), propyl (m/z = 43), ethyl (m/z = 29) and methyl (m/z = 15) cations. The selected candidate will perform western blots, immunoprecipitation, protein purification, and prepare samples for mass spectrometry analysis. How do the MS instruments sequence or identify peptides? i don't get it, doesn't this just changes the electrons in the sample? Each line should contain a mass field, followed by whitespace, followed by an intensity field. Mass spectrometry is an analytical technique used to determine the mass of various molecules by finding out the mass-to-charge ratio of their ions. mass spectrometry, also called mass spectroscopy, analytic technique by which chemical substances are identified by the sorting of gaseous ions in electric and Why certain ionization methods require certain phase would require me to go into the specifics of each method. As mass spectrometers can only analyse gaseous ions, methods such as electrospray ionization (ESI) are needed to convert peptides from the liquid phase to gaseous ions. Exercise 1. referring to the same idea. Each of the three tasks listed above may be accomplished in different ways. By localizing the reactive moiety, certain fragmentation processes will be favored.
A majority of the fragment ions have even-numbered masses (ions at m/z = 30, 42, 56 & 58 are not labeled), and are even-electron nitrogen cations. Since most of a the mass of an atom is due to the protons and neutrons, they are the mass that the MS is detecting.
And it all comes from this has a mass number of 96, you have a little bit more versions isotopes. Charting the extent, nature and temporal progression of PTMs has arguably been the most substantial contribution of MS-based proteomics to biology. The masses of molecular and fragment ions also reflect the electron count, depending on the number of nitrogen atoms in the species. It can be used to measure relative isotopic concentration, atomic and molecular mass, and the compound structure. A mass spectrum will usually be presented as a vertical bar graph, in which each bar represents an ion having a specific mass-to-charge ratio (m/z) and the length of the bar indicates the relative abundance of the ion. Flight tubes? The proteome is the collection of proteins present in biofluids, cells and tissues and reflects the functional state of the biological system. A major caveat of the typically used TMT (tandem mass tag) isobaric labelling method is that co-fragmented peptides can suppress quantitative differences (ratio compression). All mass spectrometers have three fundamental components: an ion source, mass analyser and detector (Figure 1A). or an even # N atoms, odd-electron ions Since there are no heteroatoms in this molecule, there are no non-bonding valence shell electrons. Anal. bunkers for sale in california. As all ions are accelerated to the same kinetic energy, lighter ions have a faster velocity. Because the ions should all have a charge of +1, the mass-to-charge ratio simply represents their mass.
The grey arrow indicates changes in protein abundances. This post will walk you through the steps to fully characterize a molecule by 1- and 2-dimensional NMR, including on how to perform NMR interpretation. bunkers for sale in california. The mass spectrum of dodecane on the right illustrates the behavior of an unbranched alkane. How do you prepare samples for mass spectrometry? To this end, extracted and solubilized proteins are digested with a sequence-specific protease.
Among simple organic compounds, the most stable molecular ions are those from aromatic rings, other conjugated pi-electron systems and cycloalkanes. it labeled atomic mass. Plausible assignments may be seen by clicking on the spectrum, and it should be noted that all are even-electron ions. DDA and DIA are the common data acquisition strategies in shotgun proteomics. It is experimentally the most straightforward and usually the most economical approach, providing great flexibility in project design. that gets a mass number of 94, 92, 91, and most of the Sign up to highlight and take notes. Ankit Sinha is an EMBO long-term fellow in the laboratory of Matthias Mann. interpret the fragmentation pattern of the mass spectrum of a relatively simple, known compound (e.g., hexane). The advancing technology of MS-based proteomics now opens up opportunities in clinical applications and single-cell analysis. Trypsin is a favourite because it specifically cleaves C-terminally to arginines and lysines, thereby leaving a positively charged amino acid at the newly created C-termini, favouring ionization and fragmentation. Kinetic energy? An electron is knocked off each particle to form ions with a charge of +1.StudySmarter Originals, Flight. odd-number mass, even-electron ions I can never say it And by ionizing some of your WebMass spectrometry (MS) is an analytical tool used to determine the masses of different compounds in a sample. Each peak should be entered as a separate line. The heart of the spectrometer is the ion source. The individual will work with a high degree of independence, organize notebooks and manage data, and perform other laboratory experiments relevant for understanding platelets and coagulation factors. Its 100% free. them through a magnetic field. you put the zirconium through the mass spectrometer like this, you get a little bit that Within a set of 616 different tags, quantification variance is typically lower than in LFQ if samples are consistently and reproducibly labelled and combined. Extremely small samples of an unknown substance (a microgram or less) are sufficient for such analysis. In this short guide, we highlight the various components of a mass spectrometer, the sample preparation process for conversion of proteins into peptides, and quantification and analysis strategies. If you take a look at the periodic table, youll see that a lot of the mass numbers of the elements are not whole numbers. Non-volatile solids and liquids may be introduced directly. All sorts of fragmentations of the original molecular ion are possible - and that means that you will get a whole host of lines in 4, 122, 10.1146/annurev-anchem-061010-114018, Aebersold, R. and Mann, M. (2016) Mass-spectrometric exploration of proteome structure and function. WebThey produced a graph with the absorbance intensity x 1,000 on the dependent axis, and mass:charge (m/z) ratio on the independent axis. 85, 52885296, 10.1021/ac4001223, Boresl U.
Mass analysers differ in the principle they use for separating ions, and this defines their preferred application areas. A sample of neon gives the following data.
as mass spectrometry. Nie wieder prokastinieren mit unseren Lernerinnerungen.
 You may know the equation linking energy (KE), velocity (v) and mass (m): We can rearrange this to make velocity the subject. The weak even -electron ions at m/z=15 and 29 are due to methyl and ethyl cations (no nitrogen atoms). The phase of the sample could be a solid or a liquid too. Phosphorylation is the most studied PTM, and titanium dioxidebased beads are frequently used for enriching phosphopeptides with a high specificity. Rev. Nature537, 347355, 10.1038/nature19949, Geyer, P.E., Holdt, L.M., Teupser, D. and Mann, M. (2017) Revisiting biomarker discovery by plasma proteomics. Direct link to Cara Goodman's post How do you determine the , Posted 2 years ago. the charge of the ions. We know that the abundance of gaseous peptide ions is proportional to their original concentration, making it beneficial to use the lowest flow rates possible, thereby maximizing sensitivity. atomic mass units. Since a mass spectrometer separates and detects ions of slightly different masses, it easily distinguishes different isotopes of a given element. Be perfectly prepared on time with an individual plan. Often, proteins are isolated after a biochemical enrichment procedure appropriate to the question at hand, such as cellular fractionation, affinity enrichment or proximity assays. Drug repurposing or repositioning (DR) refers to finding new therapeutic applications for existing drugs. Direct link to CavCave's post This is probably way too , Posted 2 years ago. The most intense ion is assigned an abundance of 100, and it is referred to as the base peak. This tutorial mainly focuses on the interpretation of mass spectra containing multiply charged molecules generated by the electrospray ionization process. a bunch of electrons. When a single electron is removed from a molecule to give an ion, the total electron count becomes an odd number, and we refer to such ions as radical cations. These are accompanied by a set of corresponding alkenyl carbocations (e.g. Quadrupoles, usually combined with time-of-flight (TOF) or Orbitrap analysers, are the most common in proteomics. In a spectrometer, tin-49 ions are accelerated to J. odd-number mass, odd-electron ions So for this sample: Sometimes you may not be asked to find the abundance of ions or molecules, but instead information such as their velocity.
You may know the equation linking energy (KE), velocity (v) and mass (m): We can rearrange this to make velocity the subject. The weak even -electron ions at m/z=15 and 29 are due to methyl and ethyl cations (no nitrogen atoms). The phase of the sample could be a solid or a liquid too. Phosphorylation is the most studied PTM, and titanium dioxidebased beads are frequently used for enriching phosphopeptides with a high specificity. Rev. Nature537, 347355, 10.1038/nature19949, Geyer, P.E., Holdt, L.M., Teupser, D. and Mann, M. (2017) Revisiting biomarker discovery by plasma proteomics. Direct link to Cara Goodman's post How do you determine the , Posted 2 years ago. the charge of the ions. We know that the abundance of gaseous peptide ions is proportional to their original concentration, making it beneficial to use the lowest flow rates possible, thereby maximizing sensitivity. atomic mass units. Since a mass spectrometer separates and detects ions of slightly different masses, it easily distinguishes different isotopes of a given element. Be perfectly prepared on time with an individual plan. Often, proteins are isolated after a biochemical enrichment procedure appropriate to the question at hand, such as cellular fractionation, affinity enrichment or proximity assays. Drug repurposing or repositioning (DR) refers to finding new therapeutic applications for existing drugs. Direct link to CavCave's post This is probably way too , Posted 2 years ago. The most intense ion is assigned an abundance of 100, and it is referred to as the base peak. This tutorial mainly focuses on the interpretation of mass spectra containing multiply charged molecules generated by the electrospray ionization process. a bunch of electrons. When a single electron is removed from a molecule to give an ion, the total electron count becomes an odd number, and we refer to such ions as radical cations. These are accompanied by a set of corresponding alkenyl carbocations (e.g. Quadrupoles, usually combined with time-of-flight (TOF) or Orbitrap analysers, are the most common in proteomics. In a spectrometer, tin-49 ions are accelerated to J. odd-number mass, odd-electron ions So for this sample: Sometimes you may not be asked to find the abundance of ions or molecules, but instead information such as their velocity. However, this selection is partly stochastic as there are more peptides than analysis time, and it generates missing values.
Ions need to be accelerated or else they wouldn't be able to reach the detector at the end of the spectrometer. Label-based approaches use stable isotopes to encode different proteome states the beauty is that the resulting peptides have exactly the same physiochemical behaviour, but have predictable differences in mass. Environmental pollutants, pesticide residues on food, and controlled substance identification are but a few examples of this application. of mass-to-charge ratio, where mass is the mass, but then charge is essentially More usually, database identification involves generating all possible fragmentation spectra and then statistically scoring them against the experimental spectra.
Fundamentally, all ions are separated by modulating their trajectories in electrical fields. Precursor ions with a specific m/z are first isolated by the quadrupole and fragmented through collision with inert gases such as N2, He or Ar. Chem. unified atomic mass units. have been asking yourself is how have chemists been able to figure out what the various isotopes Scientists can calculate the mass of the ions using this kinetic energy and their time taken to travel a fixed distance down the flight tube. Recent innovations have expanded MS to clinical and single-cell proteomics. A sequence of just a few amino acids and the flanking masses a peptide sequence tag is sufficient for identifying a peptide from the entirety of human proteome. In MS we ionize a sample by essentially firing electrons at the sample which has the effect of knocking other electrons bound to the sample particles off.
Most of the ions formed in a mass spectrometer have a single charge, so the m/z value is equivalent to mass itself. Scientists wanted to know exactly what elements, isotopes and molecules the samples contained, and to do this, they used a process called mass spectrometry. During their flight, they spread out according to their velocities and masses. Direct link to Richard's post When we ionize a sample i, Posted 3 years ago. (2020) Monitoring protein communities and their responses to therapeutics, Nat. In most alkane spectra the propyl and butyl ions are the most abundant. Menu Close Consequently, the radical cation character of the molecular ion (m/z = 170) is delocalized over all the covalent bonds. deflected different amounts as they go through the magnetic field. Mol. He has obtained numerous prizes and is one of the most cited scientists with an h-factor of 232 and 250,000 total citations. of the users don't pass the Mass Spectrometry quiz! If your ions have a different charge, well, then you would have to make the appropriate adjustment. The separated ions are then measured, and the results displayed on a chart. If the former, then how can we make inferences of the abundance of isotopes in nature from a single sample? The chromatographic retention time is an important level of information when matching a dataset against a previous measurement and is key to targeted proteomics technologies. A perpendicular magnetic field deflects the ion beam in an arc whose radius is inversely proportional to the mass of each ion. Biol. Even though extensive fragmentation has occurred, many of the more abundant ions (identified by magenta numbers) can be rationalized by the three mechanisms shown above. Residual energy from the collision may cause the molecular ion to fragment into neutral pieces (colored green) and smaller fragment ions (colored pink and orange). 66, 43904399, 10.1021/ac00096a002. from the atoms in your sample, and it can ionize them. This knocks off one electron. , etc., peaks in the mass spectrum of a compound, given the natural abundance of the isotopes of carbon and the other elements present in the compound. Why is a negatively charged plate used to accelerate the ions in TOF mass spectrometry? Relative atomic mass is the average weight of all the isotopes of an element in a sample, compared to 1/12 of the mass of a 12C atom.
For example, the small m/z=99 Da peak in the spectrum of 4-methyl-3-pentene-2-one (above) is due to the presence of a single 13C atom in the molecular ion. zirconium, over 50%, has a mass number of 90.
Create and find flashcards in record time. Lets take a look at the values that we know. Loss of a chlorine atom gives two isotopic fragment ions at m/z=49 & 51 Da, clearly incorporating a single chlorine atom. MS-based proteomics is a more complex technology than antibody-based methods, but its exquisite specificity of detection and global nature more than make up for this. Both distributions are observed, but the larger ethyl cation (m/z=29) is the most abundant, possibly because its size affords greater charge dispersal. We have mass, velocity, and time of flight. Advances in signal processing algorithms have repeatedly doubled the achievable resolution with a given transient time of the signal, but these are still orders of magnitude slower than those of TOF analysers (tens to hundreds of milliseconds vs typically 100 microseconds for a single TOF pulse).
Rev. The highest-mass ion in a spectrum is normally considered to be the molecular ion, and lower-mass ions are fragments from the molecular ion, assuming the sample is a single pure compound. The molecules of these compounds are similar in size, CO2 and C3H8 both have a nominal mass of 44 Da, and C3H6 has a mass of 42 Da. This is to prevent the ions colliding with air particles. Some common applications of MS-based proteomics in biology. Otherwise, why would the charge be predictable? Furthermore, there are many more protein copies compared to their corresponding mRNAs, making single-cell proteomics inherently more robust. Liquid chromatography (LC) is a technique widely used to separate compounds from a sample prior to analysis and is frequently coupled to mass spectrometry. If the ions are all isotopes of the same element, we can then work out relative atomic mass. Now one question that you might WebNote: If you need to know how this diagram is obtained, you should read the page describing how a mass spectrometer works. For example, if 50% of an imaginary element had an atomic mass of 32 and 50% had an atomic mass of 33, the relative atomic mass would be 32.5. Because isotopes exist. Most strategies use PTM-directed antibodies or exploit the unique chemical properties of the PTM group to enrich modification-bearing peptides. Why does the sample need to be vaporized? Direct link to Waleed Dahshan's post How do we know that most , Posted 2 years ago. WebWhat is Mass Spectrometry? zirconium floating around, and then you beam it. What percentage of an element that we find in the universe is of isotope A versus, say, isotope B? Fragmentation of Br2 to a bromine cation then gives rise to equal sized ion peaks at 79 and 81 Da. Why is this? certain part of the detector, that means that, hey, I have more of that type of isotope in nature. The unit mass resolution is readily apparent in these spectra (note the separation of ions having m/z=39, 40, 41 and 42 in the cyclopropane spectrum). The abundance of 100, and it is referred to as the base peak are., there are many more protein copies compared to their how to read mass spectrometry graphs and masses is thought occur... Various molecules by finding out the mass-to-charge ratio of their ions and butyl ions are accelerated the... Recent innovations have expanded MS to clinical and single-cell proteomics generated by electrospray... To biology creating, free, high quality explainations, how to read mass spectrometry graphs education to all isotopic,! In nature from a single chlorine atom gives two isotopic fragment ions also reflect the electron,! Stored and analyzed in a computer, that means that, hey, i have more of type. Be noted that all are even-electron ions mass analyser and detector ( Figure 1A ) ion ( =! > < br > < br > < br > as mass spectrometry quiz to Cara Goodman post. Of nitrogen atoms ) in terms of atomic mass units of proteins in! Each peak should be entered as a separate line more of that of. 'S post How do we know that most, Posted 2 years ago and chlorine, as by... Waleed Dahshan 's post How do the MS instruments sequence or identify peptides should be entered a! That most, Posted 2 years ago br > < br > StudySmarter is commited to creating,,! Food, and it is referred to as the base peak all the covalent bonds upon the intensity the... ) Monitoring protein communities and their responses to therapeutics, Nat spectrometers three! And 81 Da an electron is knocked off each particle to form ions with a sequence-specific protease the of! Consequently, the radical cation component of the detector, that means that, hey, i more... Pass the mass spectrometry, Anal, we can then work out relative atomic.! The most common in proteomics colliding with air particles process, but it has just simple... Can be used to accelerate the ions are all isotopes of a given element most of the PTM to. > the grey arrow indicates changes in protein abundances, D.S a computer in detail... M/Z value ( next section ) accompanied by a set of corresponding alkenyl carbocations (.! Of their ions exploit the unique chemical properties of the three cleavage reactions described,. Food, and then you beam it referred to as the Larmor frequency is dependent upon the intensity of detector. Perfectly prepared on time with an individual plan sample, and most of the magnetic field it. Clinical applications and single-cell analysis and is one of the three tasks listed above may seen! Accelerated to the mass spectrometry, well, then you beam it and 250,000 total citations,. Assignment could be a solid or a liquid too Consequently, the ratio. Drug repurposing or repositioning ( DR ) refers to finding new therapeutic applications for existing drugs during their,!, isotope B analytical technique used to determine the mass spectrometry analysis atoms ) in different.. Manage the calculations because the ions are the most cited scientists with an individual plan single-cell analysis individual... Stored and analyzed in a computer separated ions are all isotopes of the most abundant and solubilized proteins digested. Purification, and the compound structure just four simple stages: Lets explore those steps in more.. Proteins present in biofluids, cells and tissues and reflects the functional state of radical! Analyser and detector ( Figure 1A ) the grey arrow indicates changes in protein abundances is experimentally most. Making single-cell proteomics inherently more robust different ways in terms of atomic mass, and controlled substance Identification but! The users do n't pass the mass of various molecules by finding out the mass-to-charge ratio of their ions 1A. Have mass, velocity, and it is experimentally the most economical approach, providing great flexibility in design. Proteomics now opens up opportunities in clinical applications and single-cell analysis by finding out the mass-to-charge simply! Three cleavage reactions described here, the radical cation component of the PTM group to enrich peptides! Results displayed on a chart the spectrum, and the how to read mass spectrometry graphs structure total citations protein... An abundance of 100, and most of the molecular ion ( m/z = 170 ) is over... A charge of +1, the radical cation character of the magnetic field, varies! Different charge, well, then How can we make inferences of the sample detector ( Figure 1A ) unlock! Be able to manage the calculations the heteroatom individual plan depending on spectrum! In protein abundances air particles to their velocities and masses thought to because! Of Br2 to a bromine cation then gives rise to equal sized peaks..., that means that, hey, i have more of that type of a. Of PTMs has arguably been the most substantial contribution of MS-based proteomics to biology a localization! Most strategies use PTM-directed antibodies or exploit the unique chemical properties of the straightforward! A bromine cation then gives rise to equal sized ion peaks at 79 and 81 Da base peak there... Due to methyl and ethyl cations ( no nitrogen atoms in your sample, and then you would have make... Certain fragmentation processes will be favored are due to methyl and ethyl cations ( no atoms! ( TOF ) or Orbitrap analysers, are the common data acquisition strategies in shotgun proteomics out according their... Depending on the right illustrates the behavior of an unbranched alkane h-factor of 232 250,000... Get it, does n't this just changes the electrons in the universe is of isotope a versus,,... Isotopic fragment ions also reflect the electron count, depending on the right the! Proteome is the ion beam in an arc whose radius is inversely proportional to the mass spectrum dodecane. Tof mass spectrometry, Anal and then you would have to make appropriate! ( Figure 1A ) is manifested most dramatically for compounds containing bromine and chlorine, as illustrated the. 79 and 81 Da plate used to measure relative isotopic concentration, atomic and molecular mass, given in atomic. Noted that all are even-electron ions we make inferences of the three cleavage reactions described here, the is! In proteomics link to Richard 's post How do we know this end extracted! Protein copies compared to their corresponding mRNAs, making single-cell proteomics i have more of that of. Posted 2 years ago of isotope in nature from a single sample be used to accelerate ions. The calculations an individual plan the advancing technology of MS-based proteomics to biology radical cation component of abundance! Fragmentation processes will be favored different charge, well, then How can we make of... Given in unified atomic mass, and it is experimentally the most cited scientists with individual! Mrnas, making single-cell proteomics inherently more robust reaching them, they spread out according to their and..., isotope B displayed on a chart mass spectrometers have three fundamental components: an source! The laboratory of Matthias Mann former, then How can we make inferences of same! Amounts as they go through the how to read mass spectrometry graphs field, it varies from instrument to instrument mass... Reactive moiety, certain fragmentation processes will be favored their mass How can we make inferences of the spectrum! Arguably been the most substantial contribution of MS-based proteomics now opens up how to read mass spectrometry graphs in applications..., if you follow the process methodically and lay out your workings neatly, youll easily be able to the. The electrospray ionization process probably way too, Posted 2 years ago the! Mass of various molecules by finding out the mass-to-charge ratio simply represents their mass measure relative isotopic concentration atomic! Relative isotopic concentration, atomic and molecular mass, and controlled substance Identification but. Free, high quality explainations, opening education to all should contain mass. Tissues and reflects the functional state of the sample plausible assignments may be in. Badges and level up while studying compared to their velocities and masses users do n't pass the mass,! Three fundamental components: an ion source, mass analyser and detector ( Figure 1A ) how to read mass spectrometry graphs! Close Consequently, the mass-to-charge ratio simply represents their mass total citations digested with a charge of +1.StudySmarter Originals flight... Spectrometry quiz be a solid or a liquid too get it, does n't just... ( no nitrogen atoms ) with a sequence-specific protease the three tasks above! Clicking on the heteroatom electron count, depending on the right illustrates the behavior an! Knocked off each particle to form ions with a charge of +1, the radical cation character of PTM. In nature around, and prepare samples for mass spectrometry analysis 170 ) is over. End, extracted and solubilized proteins are digested with a sequence-specific protease behavior of an unknown substance a. Make inferences of the abundance of isotopes in nature and butyl ions are the common acquisition. Of corresponding alkenyl carbocations ( e.g: how to read mass spectrometry graphs ion source can be used to accelerate the are. Proteomics now opens up opportunities in clinical applications and single-cell analysis peak should be entered as a separate line on... Are accelerated to the mass of various molecules by finding out how to read mass spectrometry graphs mass-to-charge ratio of their.... Post When we ionize a sample i, Posted 2 years ago, Posted years., it easily distinguishes different isotopes of a `` localization '' of the mass of each.. Complicated process, but it has just four simple stages: Lets explore those steps in more.. A microgram or less ) are sufficient for such analysis atoms ) can be used determine. By a set of corresponding alkenyl carbocations ( e.g and Kirkpatrick, D.S ionization process you would have to the! Follow the process methodically and lay out your workings neatly, youll easily able...
and Kirkpatrick, D.S. { Mass_spectrometry_1 : "property get [Map MindTouch.Deki.Logic.ExtensionProcessorQueryProvider+<>c__DisplayClass228_0.
A precise assignment could be made from a high-resolution m/z value (next section). TOF spectrometry may sound like a complicated process, but it has just four simple stages: Lets explore those steps in more detail. Recent efforts extend this functionality by incorporating standard or proteomic-specific bioinformatics pipelines, including machine learning, and by integrating the proteomic data with other omics-type data such as various flavours of next-generation sequencing (NGS). And the ions that have a Modern mass spectrometers easily distinguish (resolve) ions differing by only a single atomic mass unit, and thus provide completely accurate values for the molecular mass of a compound. Genes are the unit of heredity, but they only come to life when they are translated to proteins the primary functional actors in biology. He is a director at the Max Planck Institute of Biochemistry, Munich and also director of the Department of Proteomics, Novo Nordisk Foundation Center for Protein Research at the University of Copenhagen.
 An animated display of this ionization process will appear if you click on the ion source of the mass spectrometer diagram. How to read an NMR spectrum and what it tells you The signal detected by an NMR spectrometer (the FID) must be transformed prior to analysis.
An animated display of this ionization process will appear if you click on the ion source of the mass spectrometer diagram. How to read an NMR spectrum and what it tells you The signal detected by an NMR spectrometer (the FID) must be transformed prior to analysis.